
Welcome to The DrugDiscoveryLab of the Department of Pharmacy at University of Naples, Federico II, Italy. We are interested in developing new strategies for drug design and discovery by integrating informatics, chemistry, biology, and medicine. Currently, we are using chemical and structural strategies to understand basic mechanisms of human diseases. Most of the work conducted in our group is aimed at developing and applying new computational approaches to accelerate the discovery of drug-like compounds. We have several years experience in supporting interdisciplinary medicinal chemistry programs through molecular modeling approaches including:
- ligand- and structure-based virtual screening cascade;
- machine learning techniques;
- polypharmacology approaches;
- homology modeling;
- QSAR and 3D QSAR;
- pharmacophore development;
- docking strategies;
- ab initio methods;
- classical molecular dynamics (MD) simulations coupled with first-principles-based computational methods (e.g. QM/MM simulations) and enhanced sampling techniques for free energy estimation.
With this general idea in mind, and in active collaboration with other biologists and pharmacologists, our group is dedicated to establish a strong and creative research program that applies state-of-the-art chemical approaches to biological problems impacting diagnosis, prevention and treatment of human diseases.
Drug Discovery Lab CPU-GPU Cluster is an in-house computational infrastructure that leverages the synergistic cluster coupling of CPUs, GPUs, displays, and storage. The cluster consists of the following hardware configuration:
- 4 CPU compute nodes (76 core, 150 GB RAM)
- 2 GPU compute nodes (5 GPU cards, 10k CUDA cores, fron NVIDIA Tesla K20m until NVIDIA GTX960)
- 2 storage nodes with 15 TB of capacity
- 8 user workstations
CPU computing

|
GPU Computing

|
Docking

|
ab-initio calculations

|
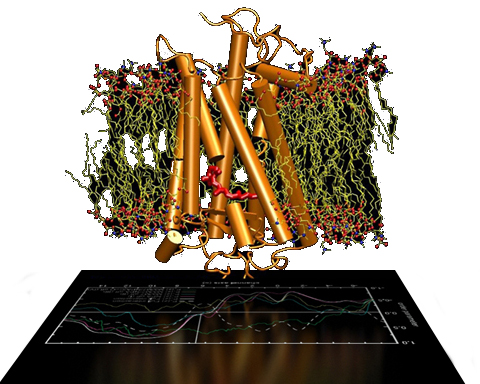
Molecular Dynamics
|
Structural Database Analysis

|


